
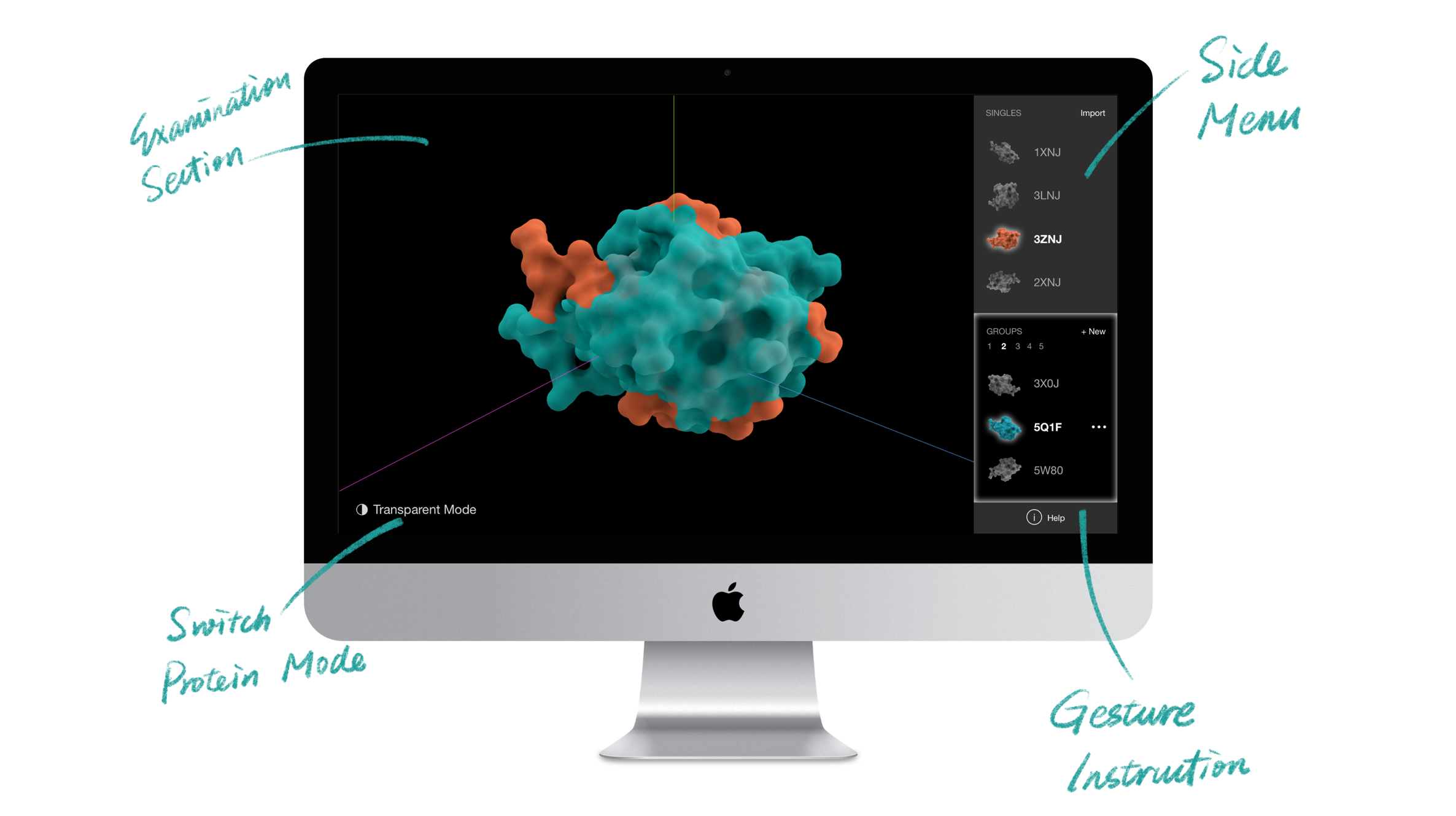
A 3D gesture-based software powered by Leap Motion Controller allows biologists without computational backgrounds to examine, compare, and classify protein surfaces with hand gestures.
There are existing tools for rendering protein structures, but the interfaces for engaging with these visualization tools are riddled with complex rendering options and analysis. They are difficult to use for investigators without computational backgrounds. Protein Renderer targets one scientific goal to visually explicate and compare binding cavities in protein structures, indicating the molecular partners that blind to a protein.
The software allows researchers to explore the protein structures by manipulating 3-D renderings via rotation and scaling. It also aids researchers in categorizing similar proteins into groups. These capabilities support the discovery and classification of protein binding sites. Classification is a novel capability not supported in existing visualization tools.
Gesture based interface is a more immersive way to engage users in content. It is popularly used in gaming and entertainment (e.g. Wii, Xbox Kinect). So, why not apply this technique in academia and science? It encourages users to interact with the objects in a more direct and natural way. This user-centered experience encourages innovation throughout research, learning, and collaboration.
This bioinformatics project was initiated by two Lehigh developers and supervised by a computer science professor. Before I joined, the two developers implemented an engineering prototype that focused on getting the functionalities to work. I was lucky to learn about the project and join the team as a UI designer to focus on improving the usability.

Protein grouping is not a familiar subject to most people. There is no existing user mental model for this task.
The previous hand gestures were only for functional purposes. They were not intuitive and users had to manually remember them to perform various functions.
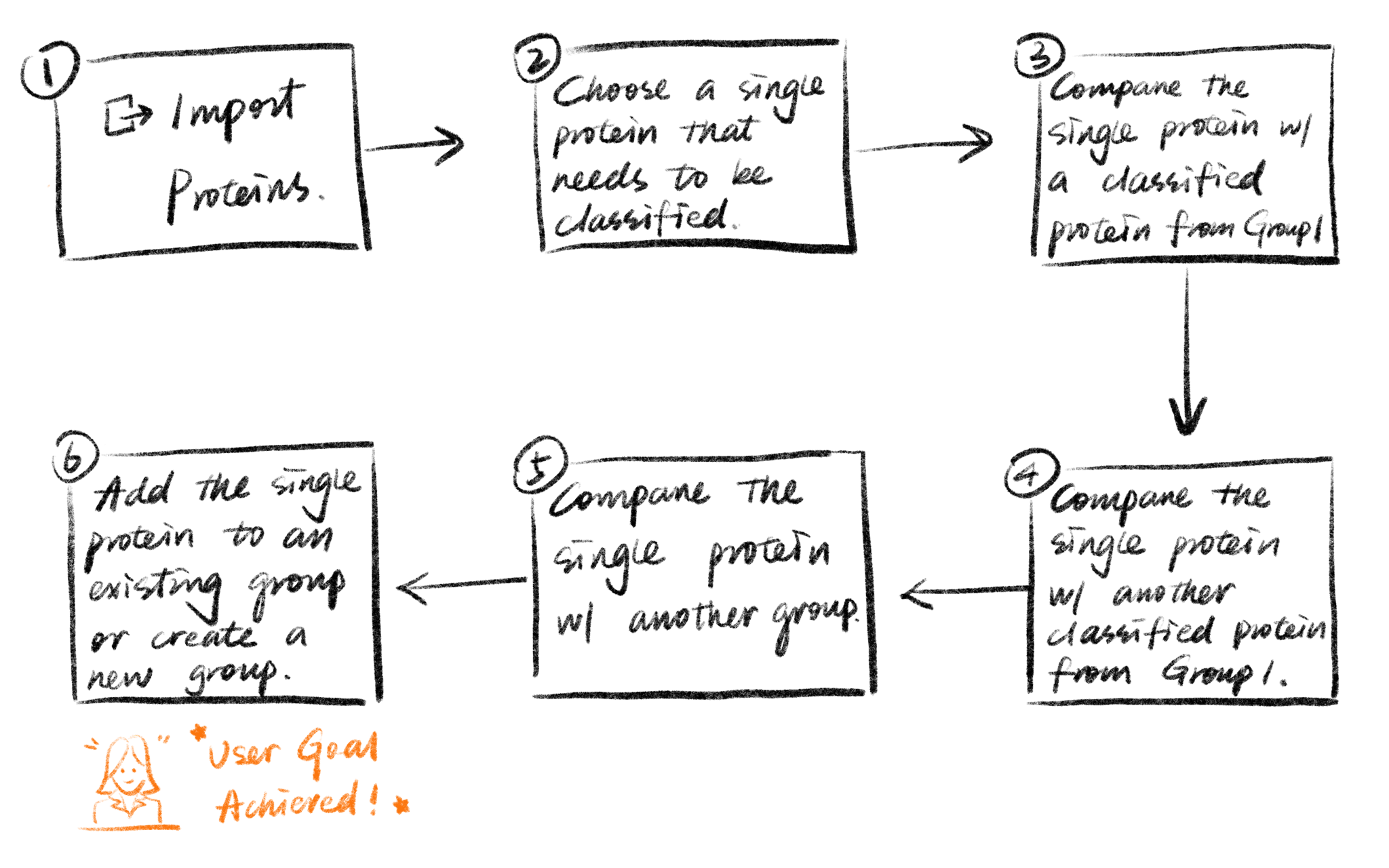
To design the layout, the first step is to discover how users expect to see the content grouped in the tool. Since protein viewing and classification are novel features, there were no established user mental models for these tasks. To build the information architecture, I decided to use the card sorting method. I put all the visual elements and functions on cards and asked the users to sort them into predefined categories or new categories.
Users were asked to pretend that they have magical powers to control everything with their hands. Then they viewed several screens and were asked to perform different tasks such as rotating the protein, putting a protein in a group, or deleting a protein. I observed and documented their gestures, then analyzed the most popular and effective gestures.
To do usability testing for a 3D gesture-based software, we had to be creative. I asked the users to complete certain tasks with gestures, and my teammate mocked the software feedback behind the scene, with a mouse.
Gestures: Go to Next/Previous Section, Expand & Collapse Side Menu
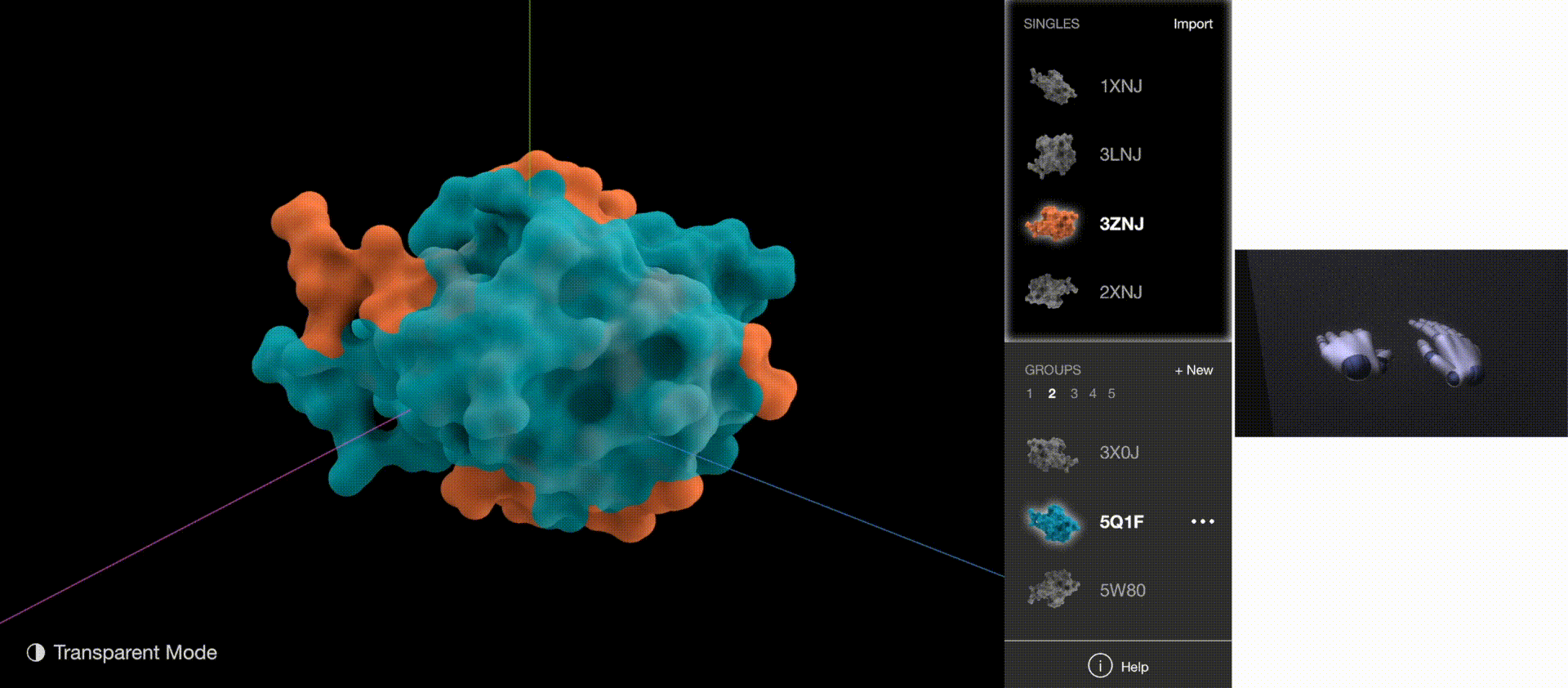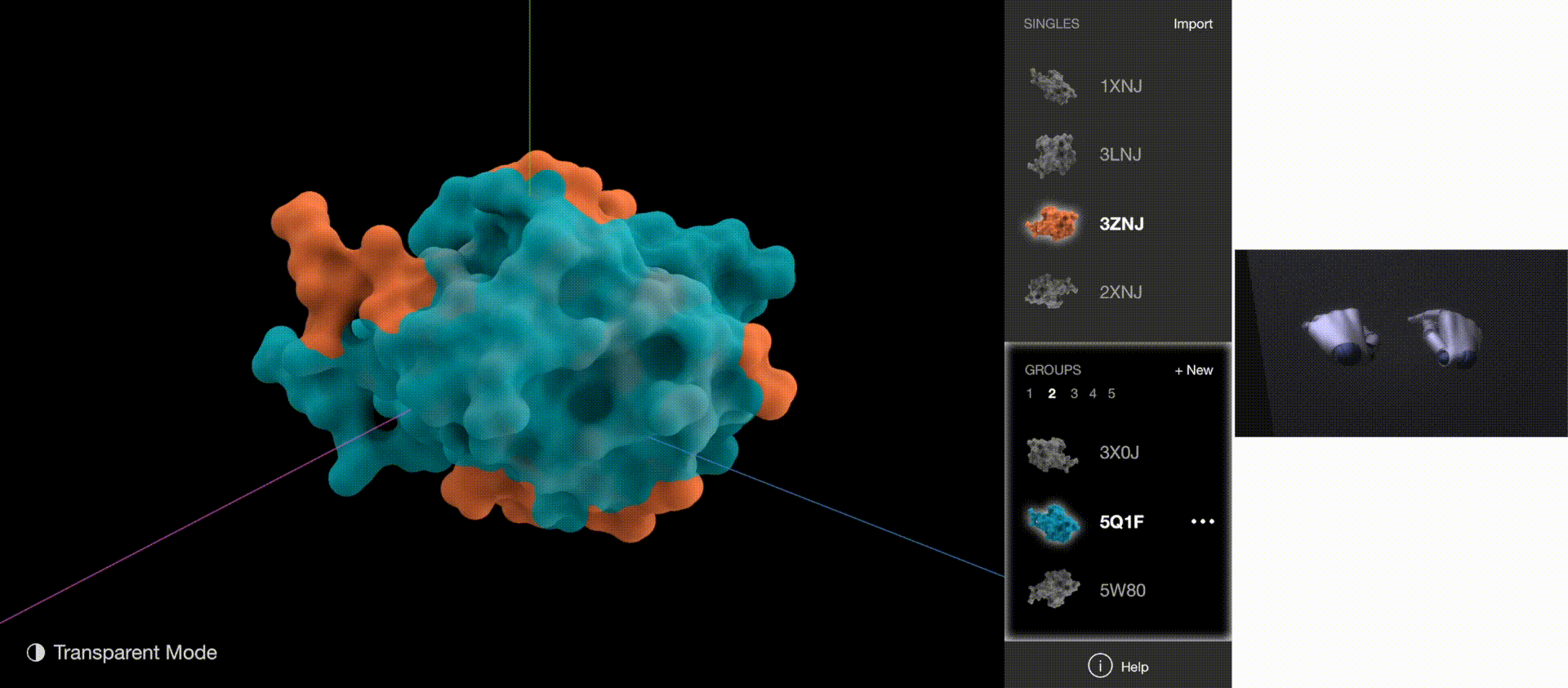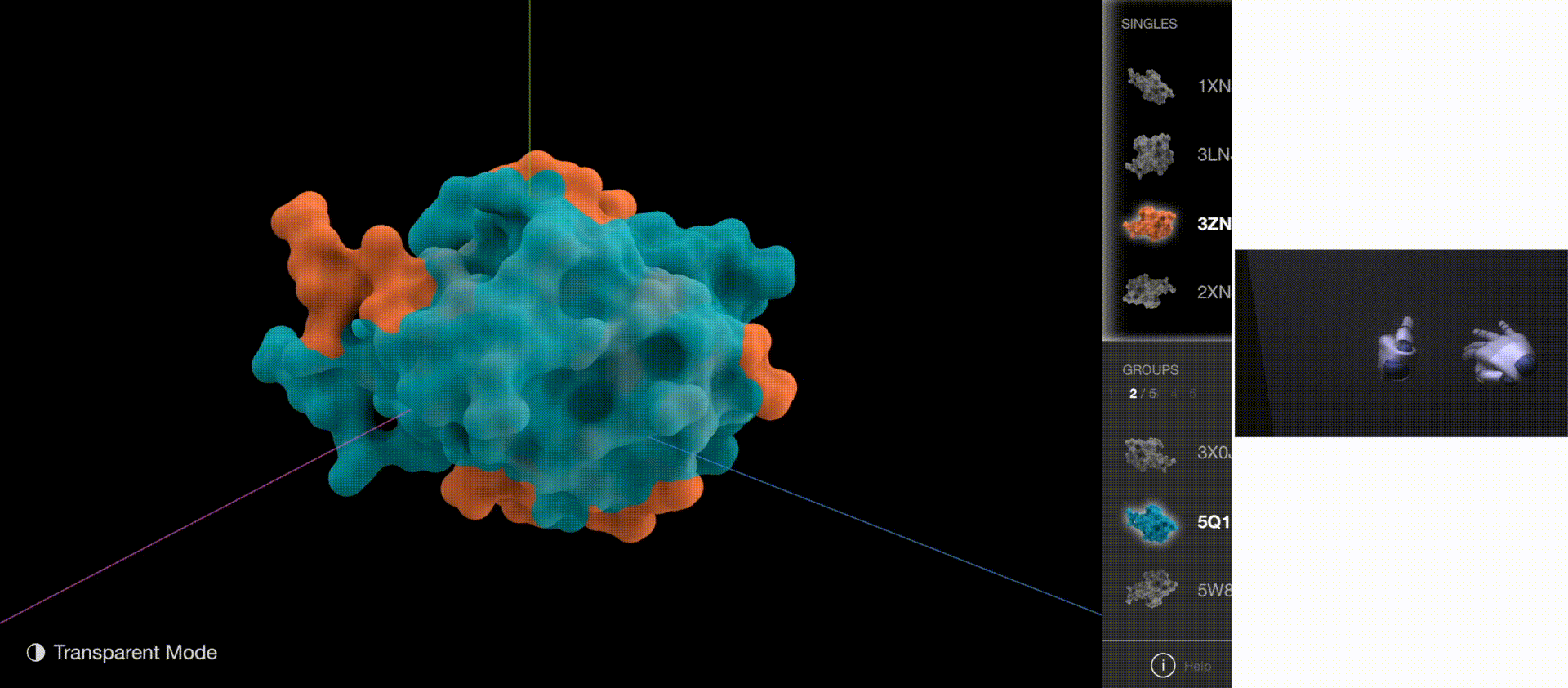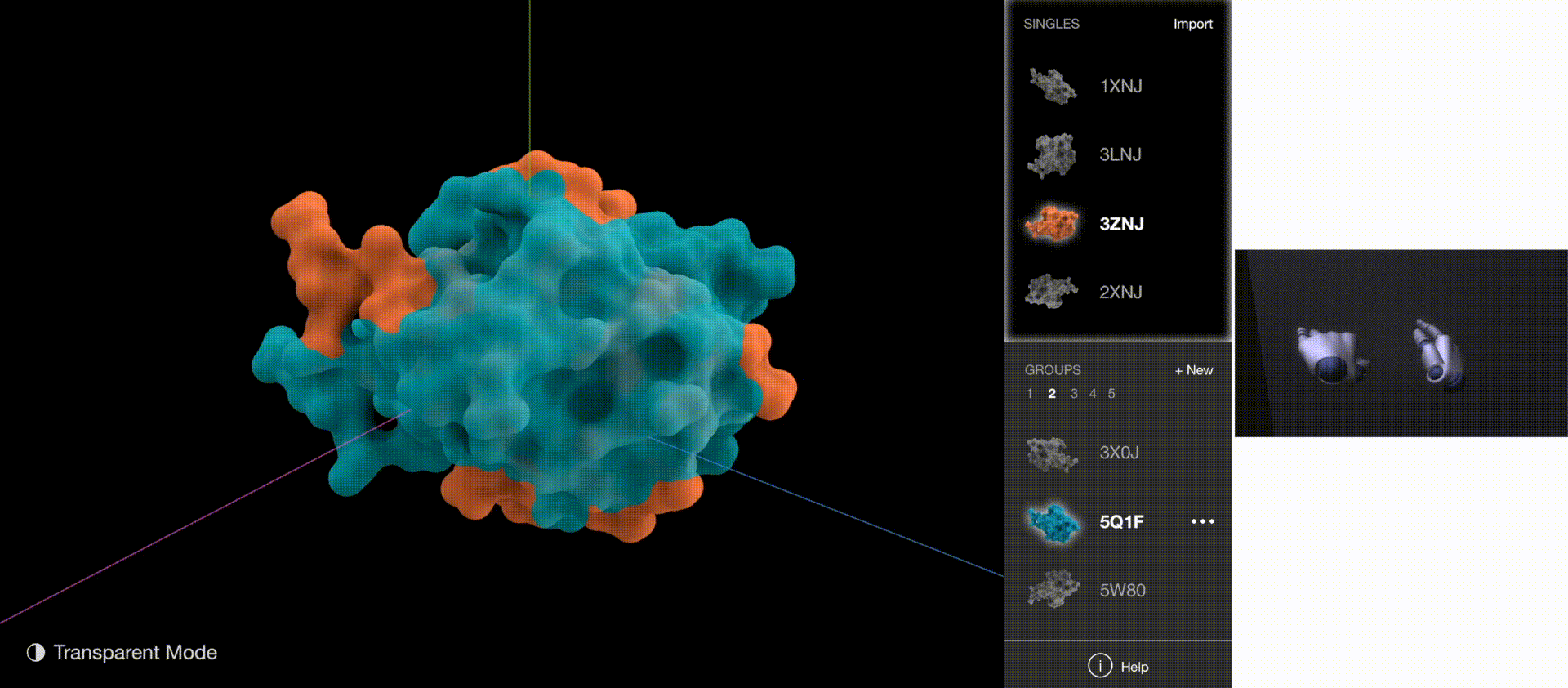
Gestures: Zoom & Rotate

Gestures: Go to Next/Previous Protein
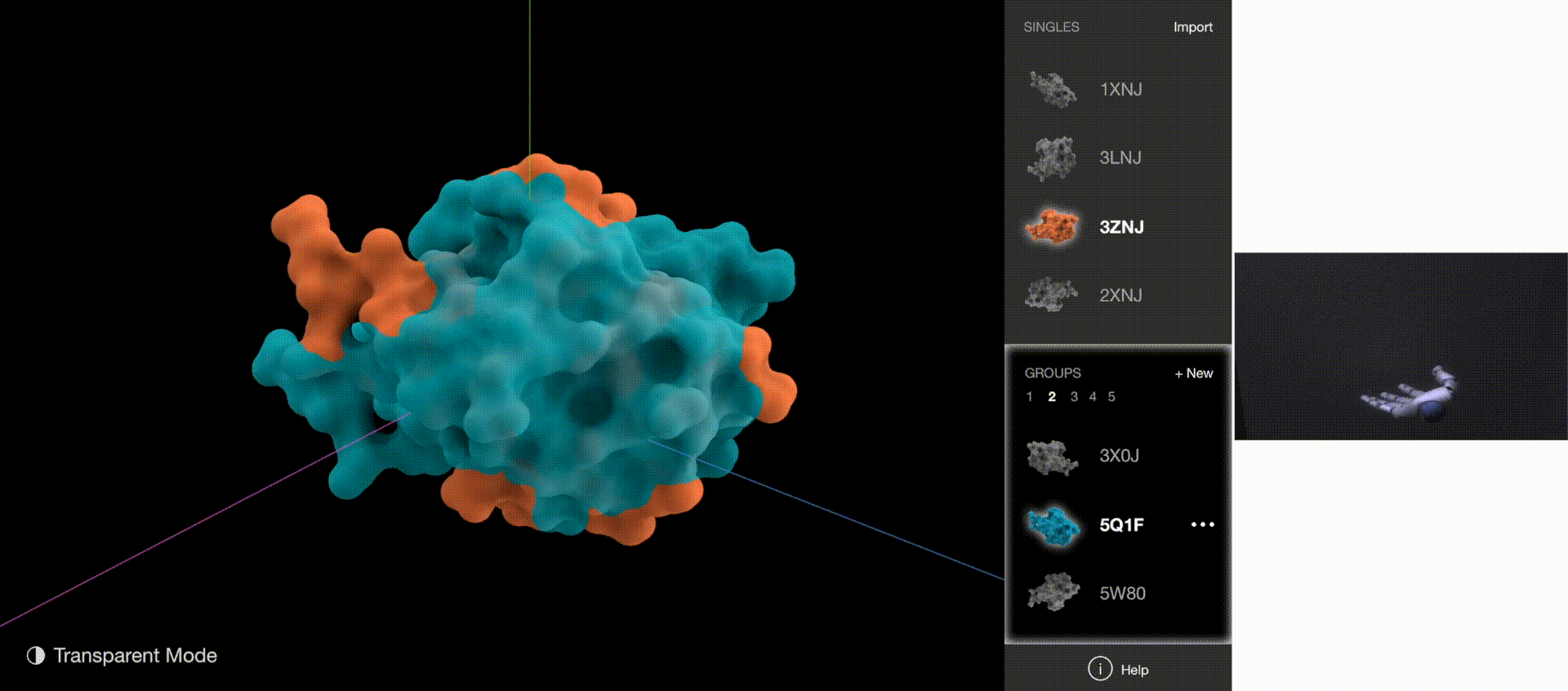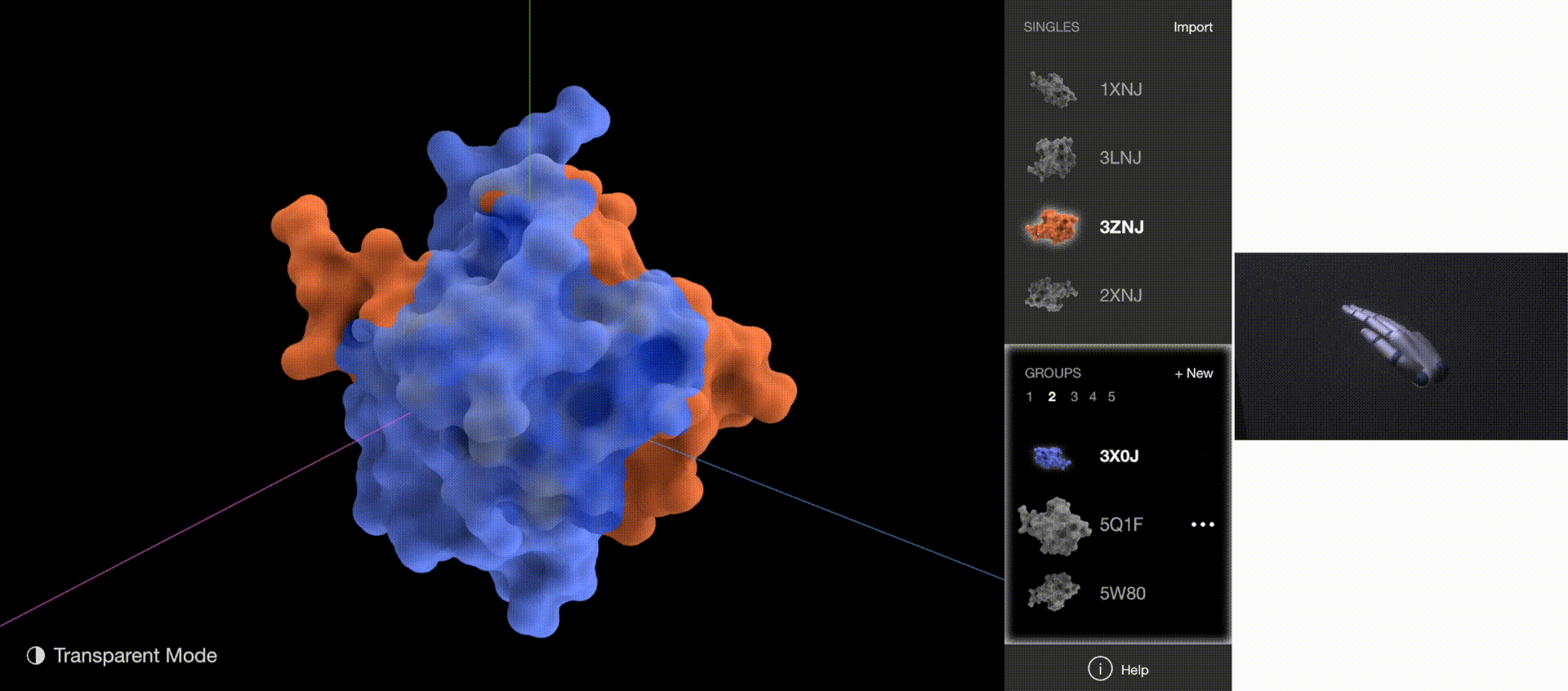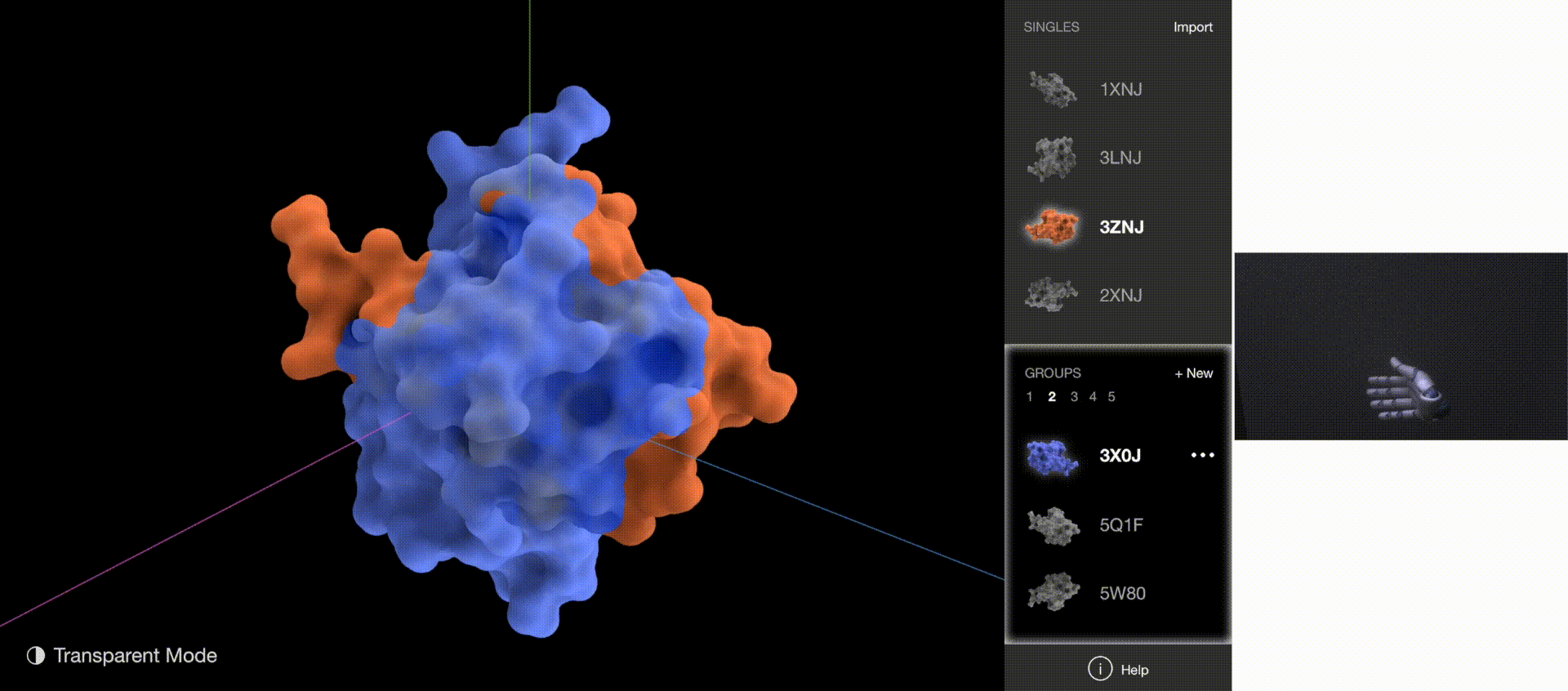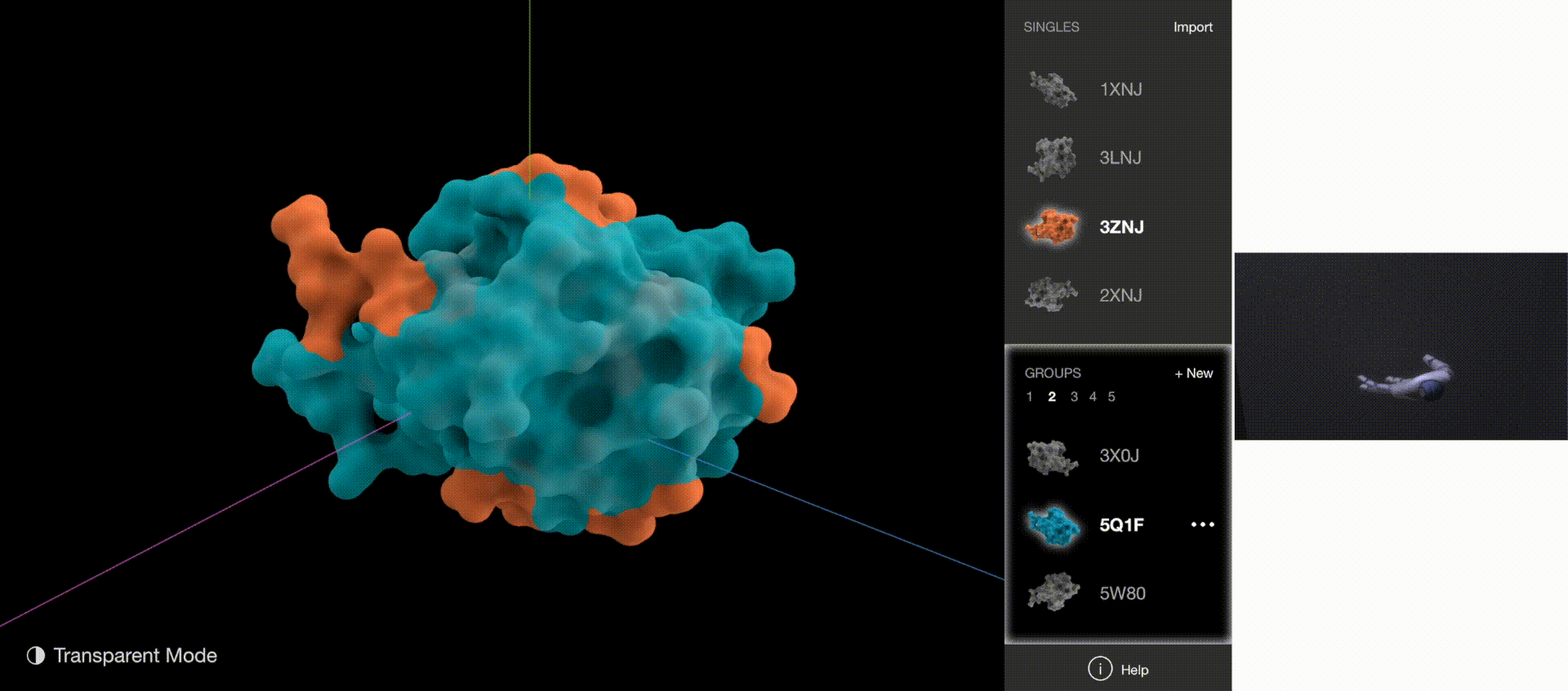
Gestures: Go to Next/Previous Group
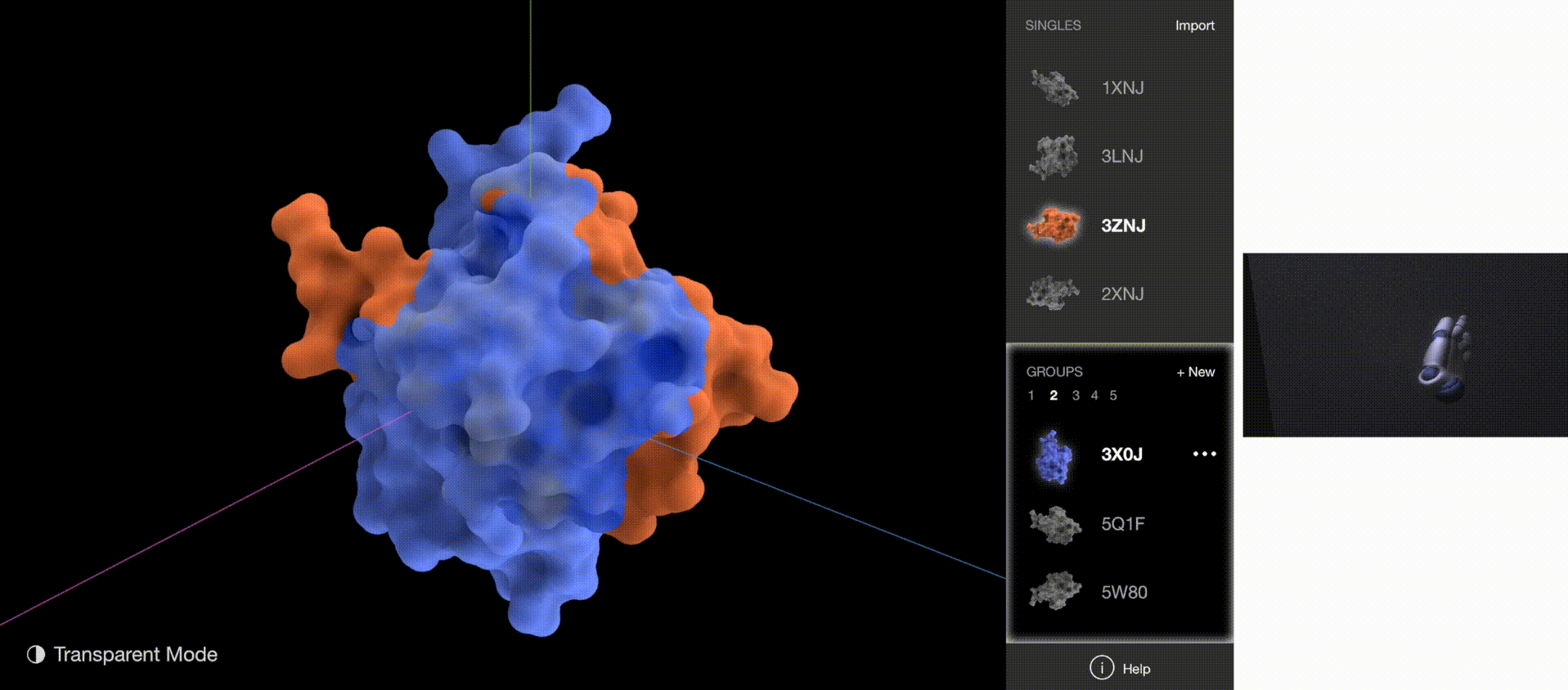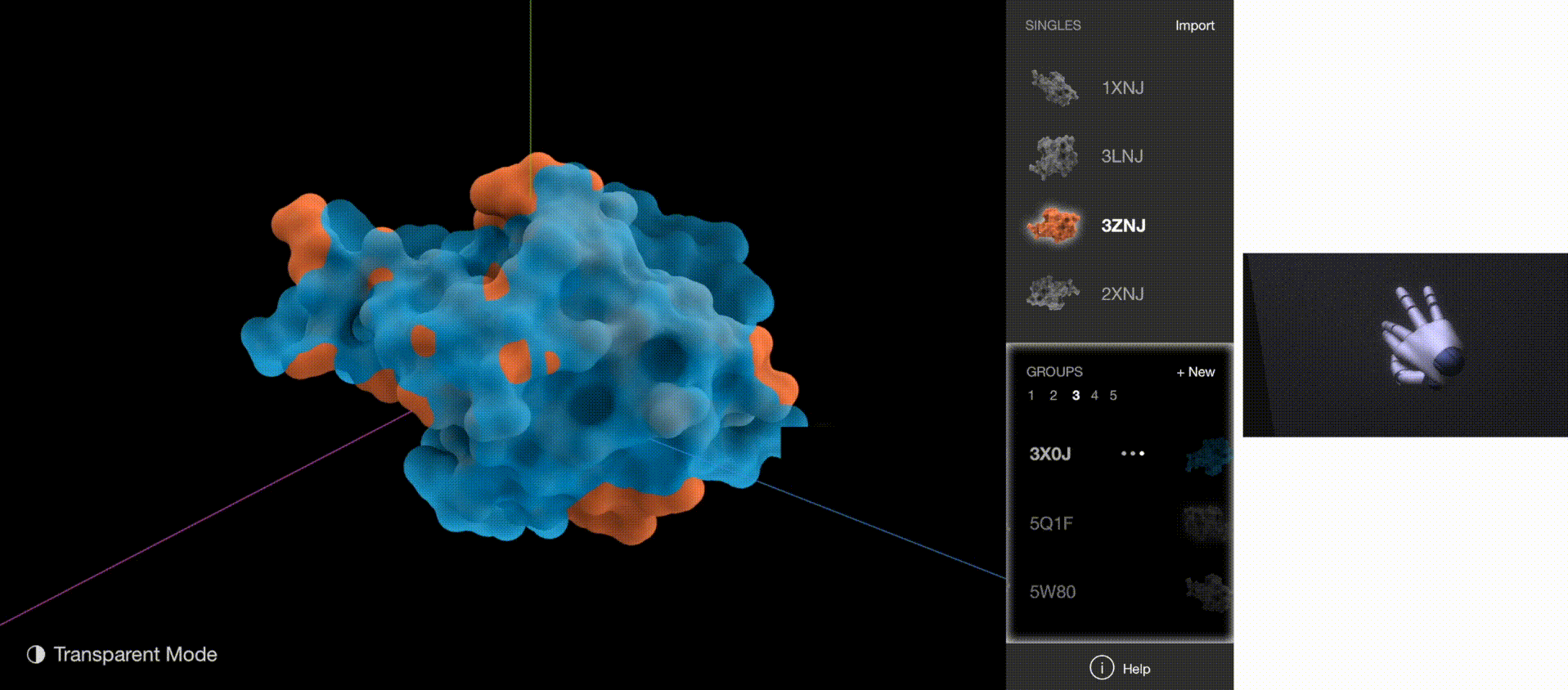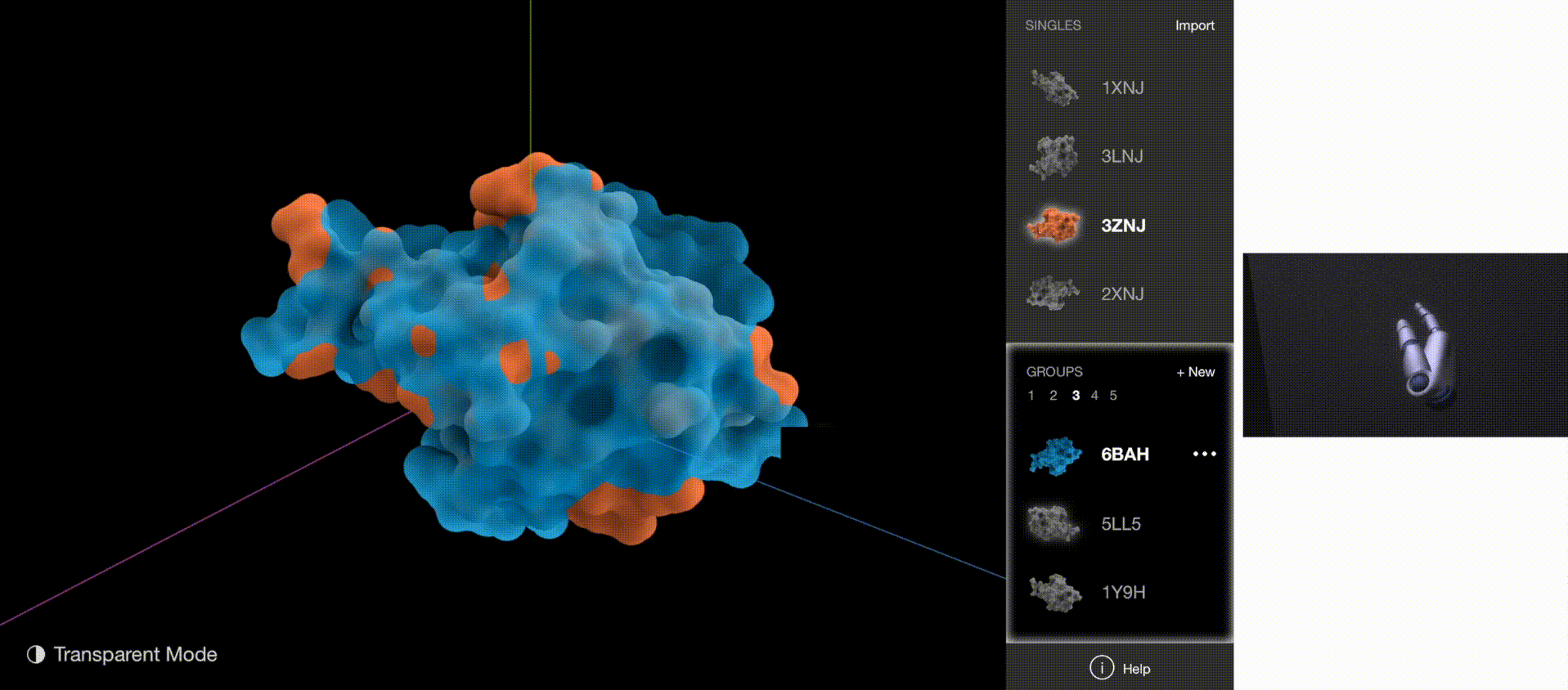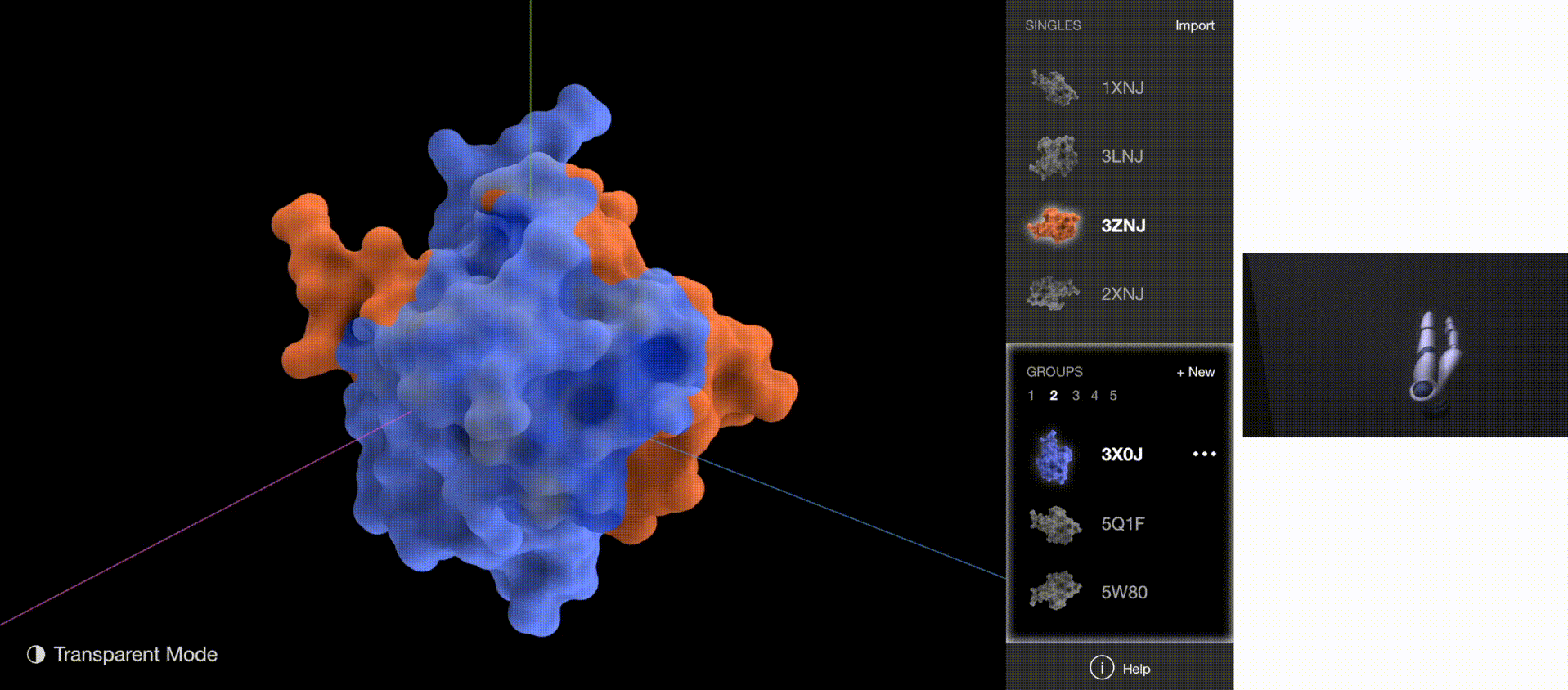
To validate whether the new gestures are more effective, I gathered quantitative data on how quickly users understood the gestures and how quickly users began implementing the correct gestures for each task. The new gestures reduced the memorization time from 5 minutes to 1 minute, and user reaction time from 5 seconds to 2 seconds.
As a team, we published a research paper. Then, we were invited to the Da Vinci Science Center to present the project.